4.3 Biplots
The details on how to create biplots is already explained in protocol 1. Concerning protocol 2, we can compare the results of version 2a (RRD, PCoA) and 2b (NI, NMDS).
4.3.1 Biplot 2D
arrows_label_adj <- rbind(c(.5,.8),c(.5,1),c(.5,1),c(.5,0),c(.5,1),
c(.5,0),c(0,.5))
row.names(arrows_label_adj) <- c("L48","L24","L5","L36","S7",
"S8","S11")
biplot2d3d::biplot_2d(prot2a_2d,
ordination_method = "PCoA",
invert_coordinates = c (TRUE,TRUE),
xlim = c(-.26,.35),
ylim = c(-.31,.35),
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_label_cex = 0.6,
arrow_mim_dist = 0.5,
arrow_label_cex = 0.6,
arrow_fig = c(.6,.95,0,.35),
arrow_label_adj_override = arrows_label_adj,
subtitle = prot2a_2d$sub2D,
test_text = prot2a_tests$text(prot2a_tests),
test_cex = 0.8,
test_fig = c(0, 0.5, 0.62, .99),
fitAnalysis_fig = c(0,.7,.05,.5),
output_type = "preview")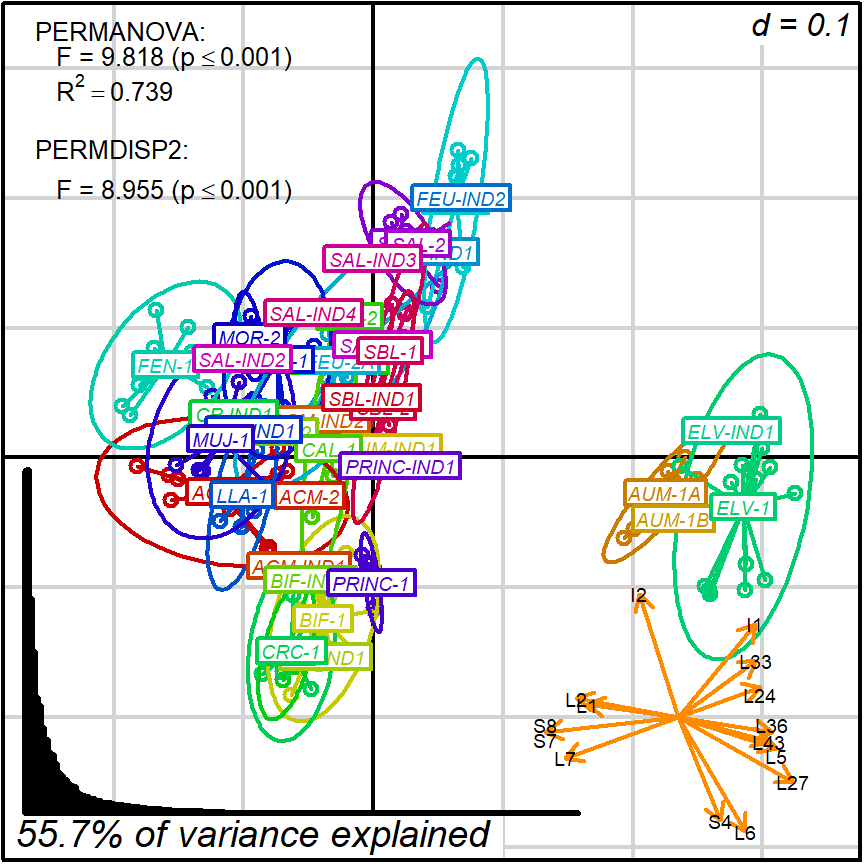
Figure 4.1: protocol 2a
arrows_label_adj <- rbind(c(.5,1),c(.5,0),c(.5,1),c(.5,1),c(.5,0),
c(0,.5),c(1,.5))
row.names(arrows_label_adj) <- c("S7","S8","CLAY","L24","L43",
"L5","L36")
biplot2d3d::biplot_2d(prot2b_2d,
ordination_method = "NMDS",
xlim = c(-.42,.38),
ylim = c(-.45,.25),
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_label_cex = 0.6,
arrow_mim_dist = .5,
arrow_label_cex = 0.6,
arrow_fig = c(.6,.95,0,.35),
arrow_label_adj_override = arrows_label_adj,
subtitle = prot2b_2d$sub2D,
test_text = prot2b_tests$text(prot2b_tests),
test_cex = 0.8,
test_fig = c(0, 0.5, 0.62, .99),
fitAnalysis_stress_axis_cex = 0.8,
fitAnalysis_fig = c(.1,.6,.1,.4),
output_type = "preview")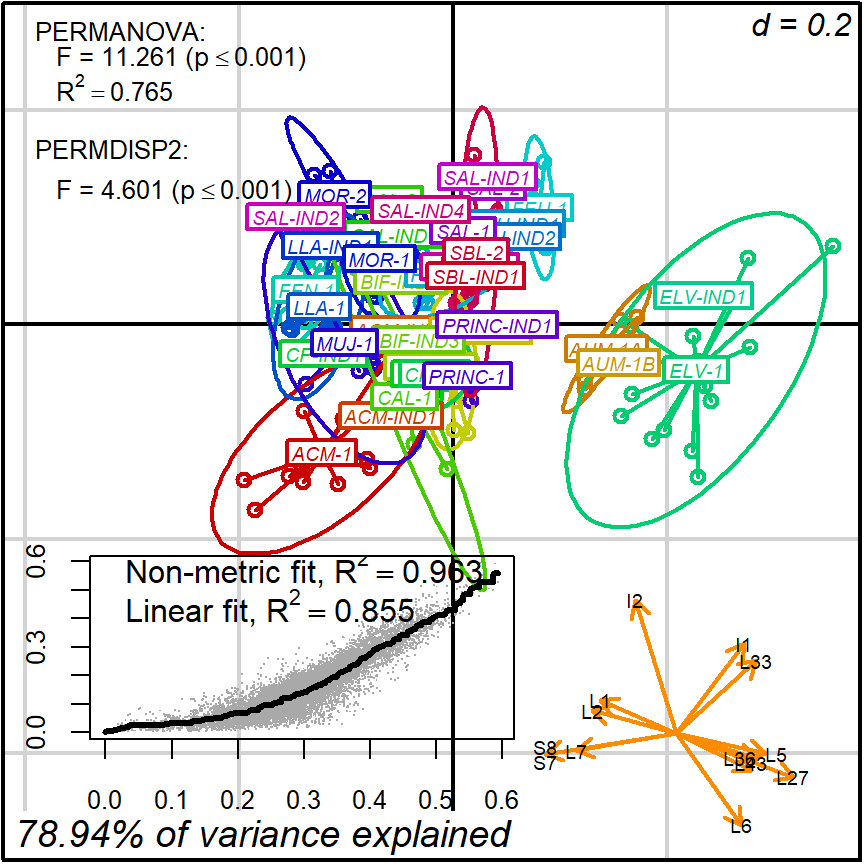
Figure 4.2: protocol 2b
4.3.2 Biplot 3D
biplot2d3d::biplot_3d(prot2a_3d,
ordination_method = "PCoA",
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_representation = "stars",
star_centroid_radius = 0,
star_label_cex = .8,
arrow_min_dist = .5,
arrow_body_length = .025,
subtitle = prot2a_3d$sub3D,
test_text = prot2a_tests$text(prot2a_tests),
test_cex = 1.25,
test_fig = c(0, 0.5, 0.65, .99),
view_zoom = 0.9)
biplot2d3d::animation(directory = directories$prot2,
file_name = "Prot2a_Biplot3D")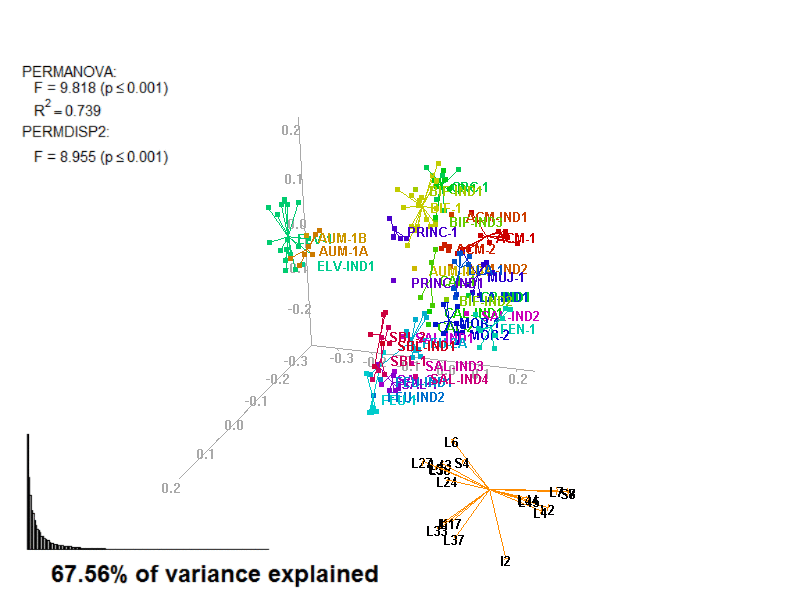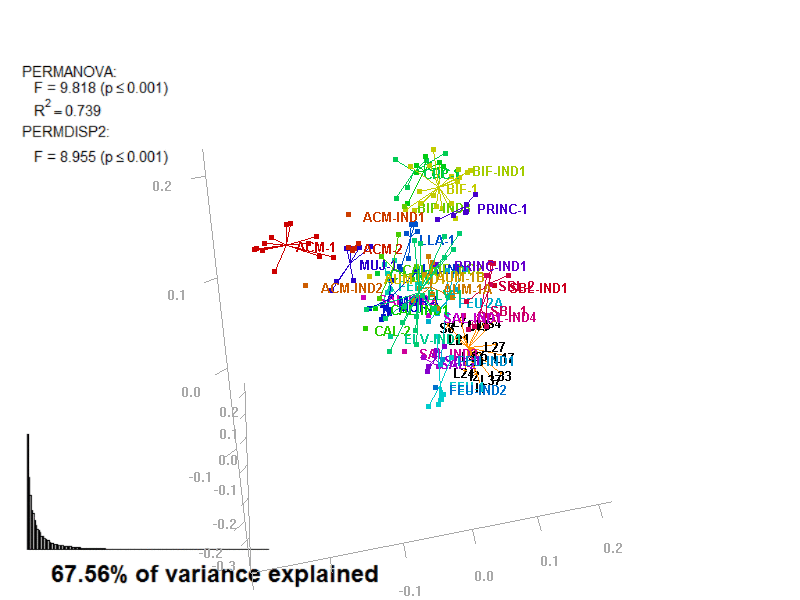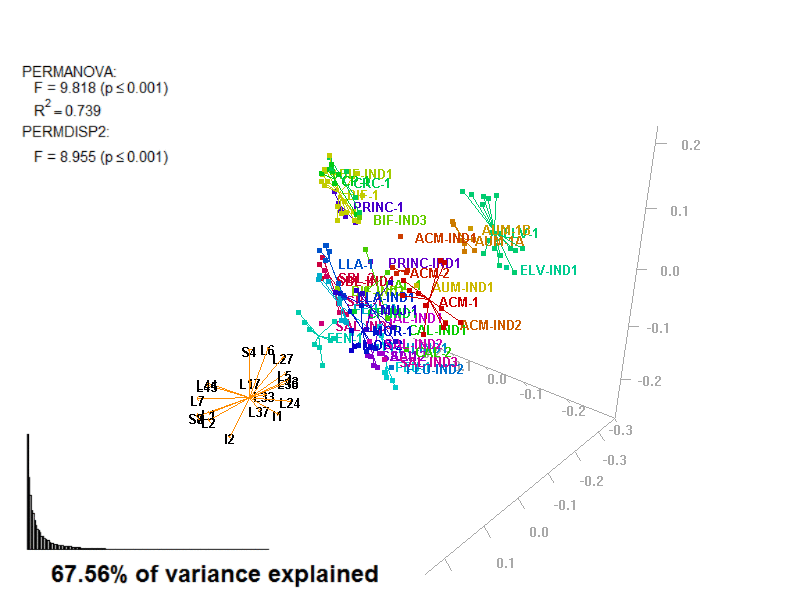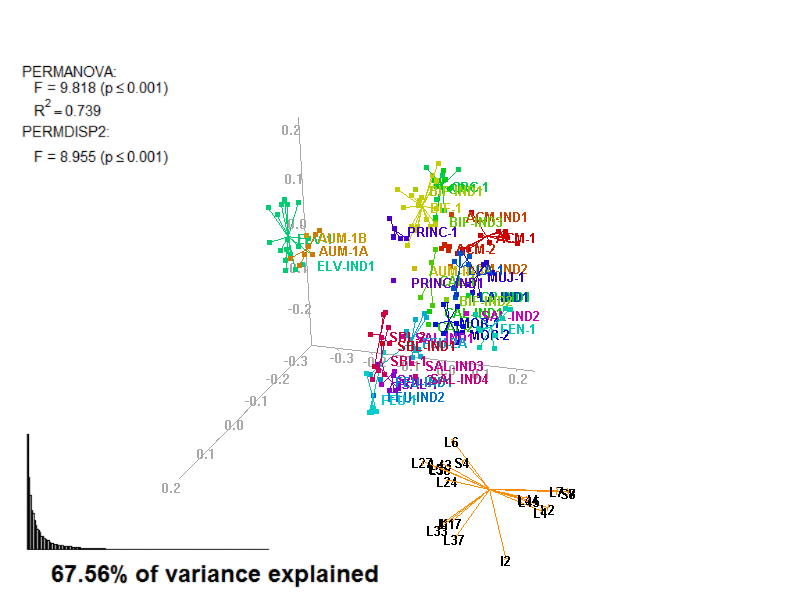
Figure 4.3: Prot2_Biplot3D.gif
NOTE: Animated GIF will not be displayed in the pdf version of this document.
biplot2d3d::biplot_3d(prot2b_3d,
ordination_method = "NMDS",
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_representation = "stars",
star_centroid_radius = 0,
star_label_cex = .8,
arrow_min_dist = .5,
arrow_body_length = .025,
subtitle = prot2b_3d$sub3D,
test_text = prot2b_tests$text(prot2b_tests),
test_cex = 1.25,
test_fig = c(0, 0.5, 0.65, .99),
view_zoom = 0.9)
biplot2d3d::animation(directory = directories$prot2,
file_name = "Prot2b_Biplot3D")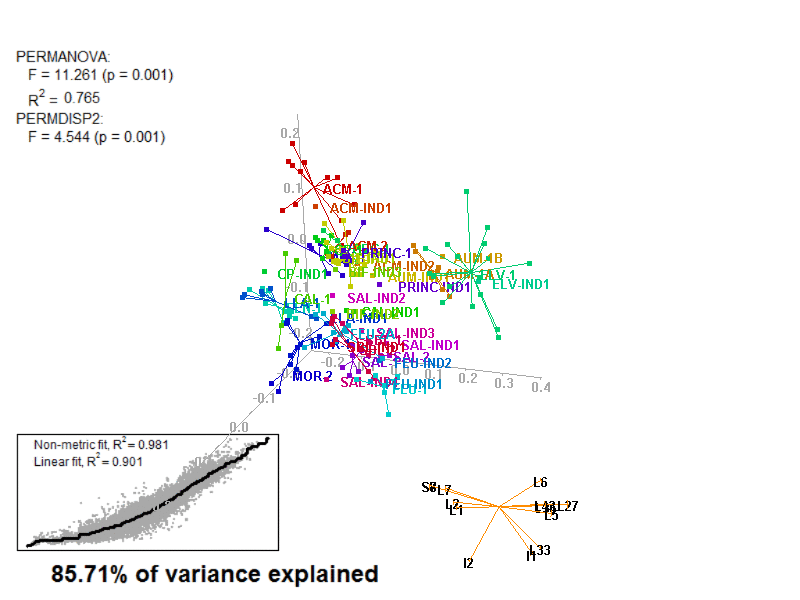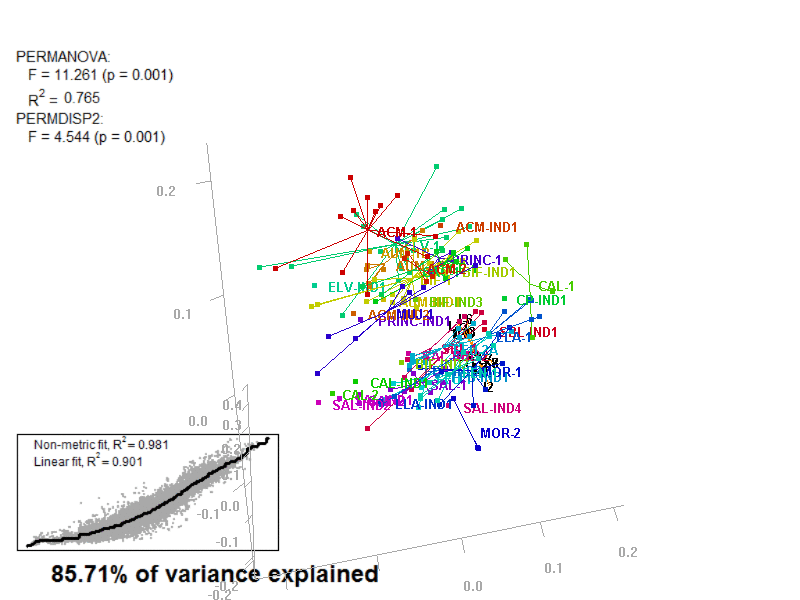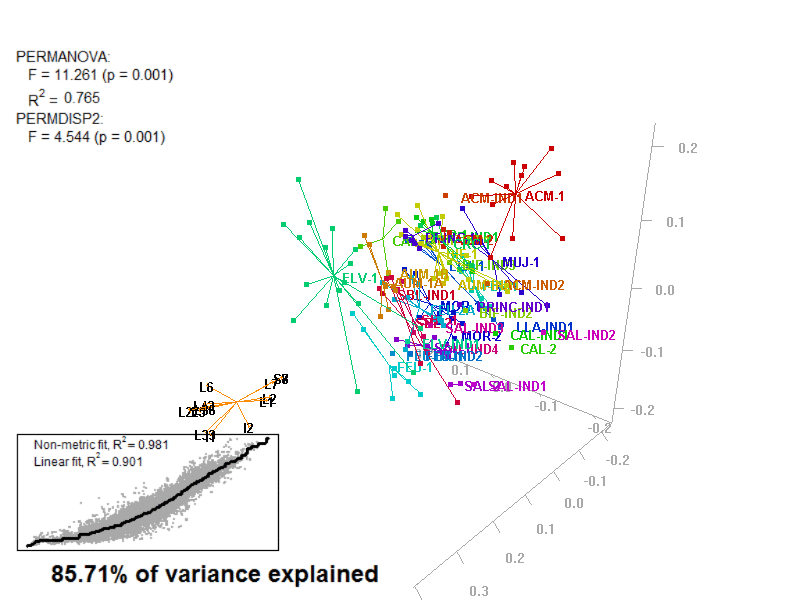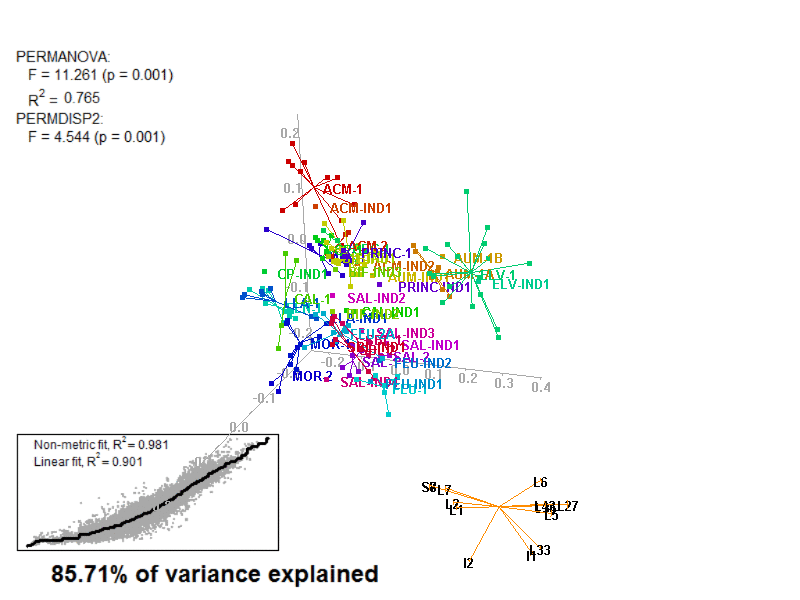
Figure 4.4: E2_Prot2b_Biplot3D.gif
NOTE: Animated GIF will not be displayed in the pdf version of this document.