4.3 Biplots
The details on how to create biplots is already explained in protocol 1. Concerning protocol 2, we can compare the results of version 2a (RRD, PCoA) and 2b (NI, NMDS).
4.3.1 Biplot 2D
arrows_label_adj <- rbind(c(.5,.8),c(.5,1),c(.5,1),c(.5,0),c(.5,1),
c(.5,0),c(0,.5))
row.names(arrows_label_adj) <- c("L48","L24","L5","L36","S7",
"S8","S11")
biplot2d3d::biplot_2d(prot2a_2d,
ordination_method = "PCoA",
invert_coordinates = c (TRUE,TRUE),
xlim = c(-.26,.35),
ylim = c(-.31,.35),
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_label_cex = 0.6,
arrow_mim_dist = 0.5,
arrow_label_cex = 0.6,
arrow_fig = c(.6,.95,0,.35),
arrow_label_adj_override = arrows_label_adj,
subtitle = prot2a_2d$sub2D,
test_text = prot2a_tests$text(prot2a_tests),
test_cex = 0.8,
test_fig = c(0, 0.5, 0.62, .99),
fitAnalysis_fig = c(0,.7,.05,.5),
output_type = "preview")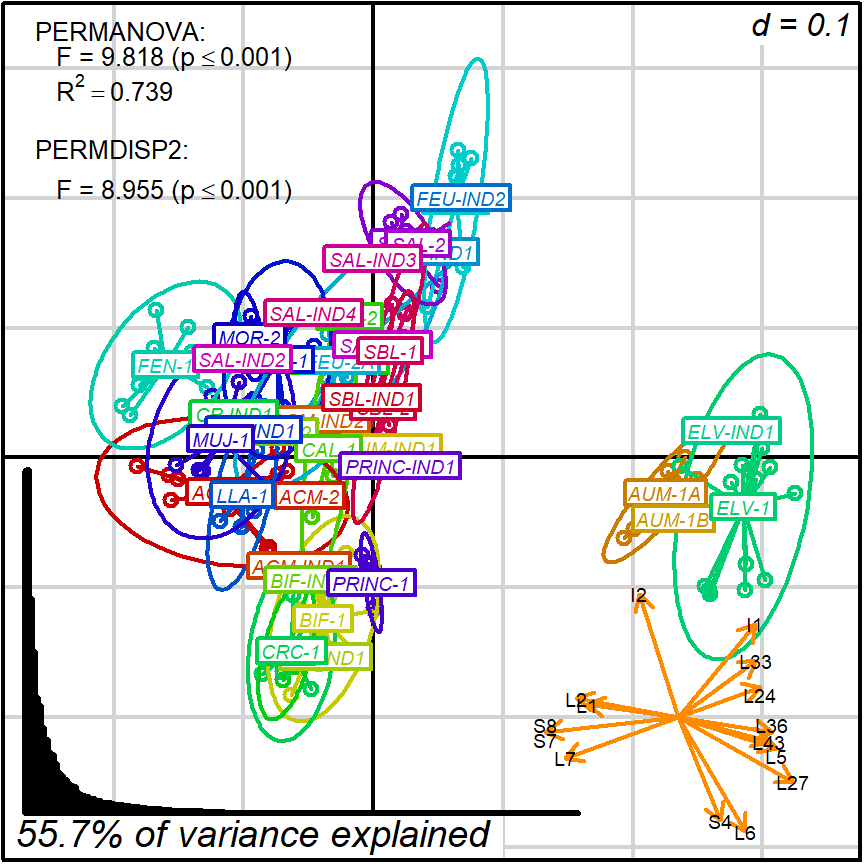
Figure 4.1: protocol 2a
arrows_label_adj <- rbind(c(.5,1),c(.5,0),c(.5,1),c(.5,1),c(.5,0),
c(0,.5),c(1,.5))
row.names(arrows_label_adj) <- c("S7","S8","CLAY","L24","L43",
"L5","L36")
biplot2d3d::biplot_2d(prot2b_2d,
ordination_method = "NMDS",
xlim = c(-.42,.38),
ylim = c(-.45,.25),
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_label_cex = 0.6,
arrow_mim_dist = .5,
arrow_label_cex = 0.6,
arrow_fig = c(.6,.95,0,.35),
arrow_label_adj_override = arrows_label_adj,
subtitle = prot2b_2d$sub2D,
test_text = prot2b_tests$text(prot2b_tests),
test_cex = 0.8,
test_fig = c(0, 0.5, 0.62, .99),
fitAnalysis_stress_axis_cex = 0.8,
fitAnalysis_fig = c(.1,.6,.1,.4),
output_type = "preview")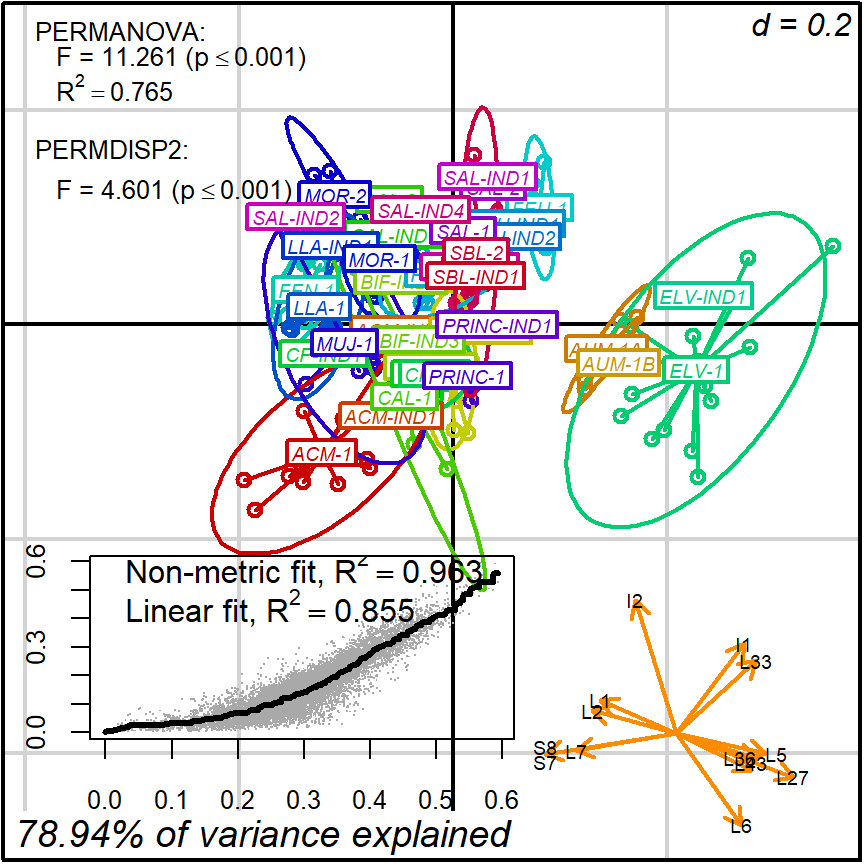
Figure 4.2: protocol 2b
4.3.2 Biplot 3D
biplot2d3d::biplot_3d(prot2a_3d,
ordination_method = "PCoA",
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_representation = "stars",
star_centroid_radius = 0,
star_label_cex = .8,
arrow_min_dist = .5,
arrow_body_length = .025,
subtitle = prot2a_3d$sub3D,
test_text = prot2a_tests$text(prot2a_tests),
test_cex = 1.25,
test_fig = c(0, 0.5, 0.65, .99),
view_zoom = 0.9)
biplot2d3d::animation(directory = directories$prot2,
file_name = "Prot2a_Biplot3D")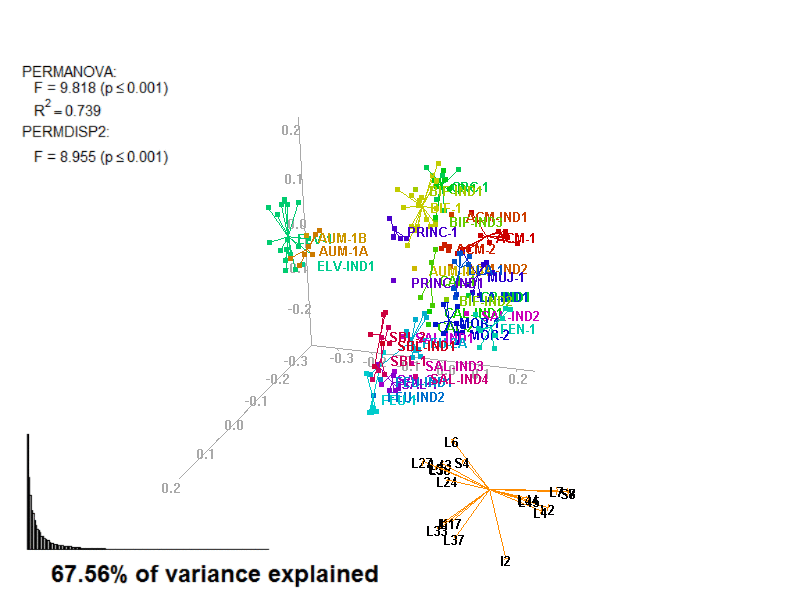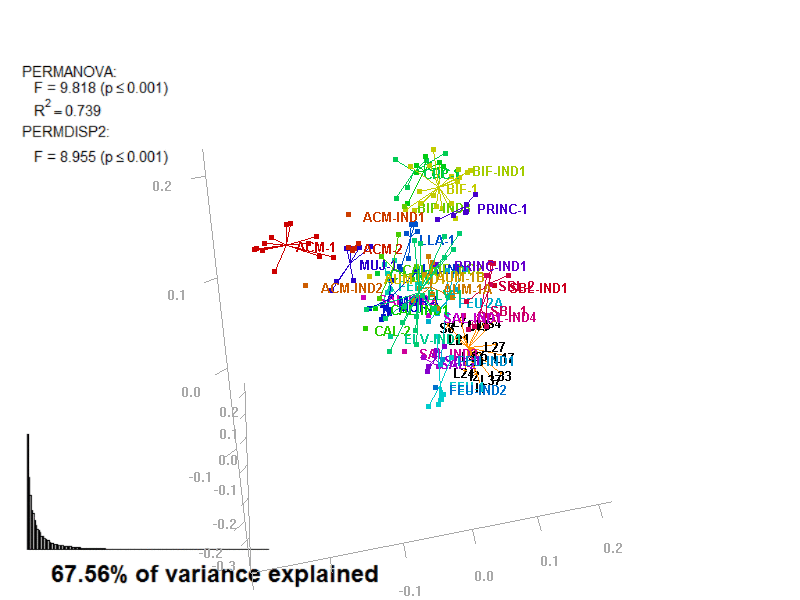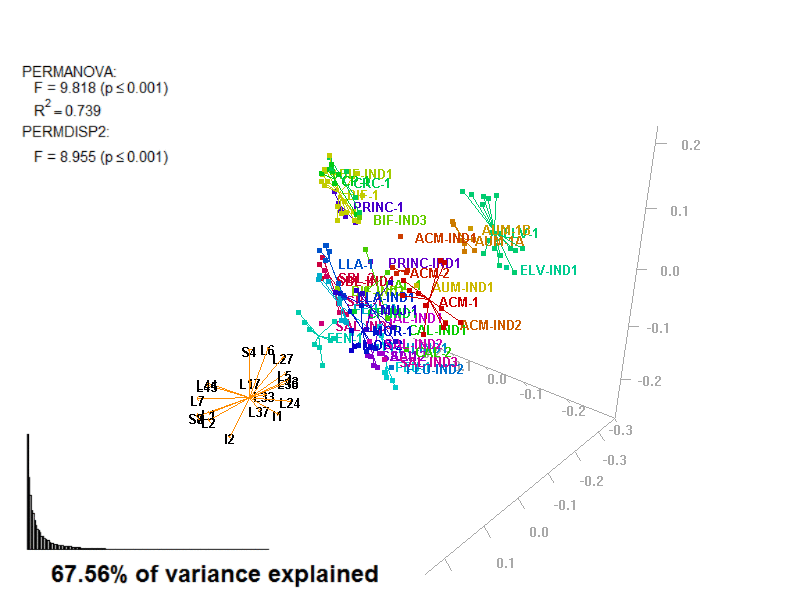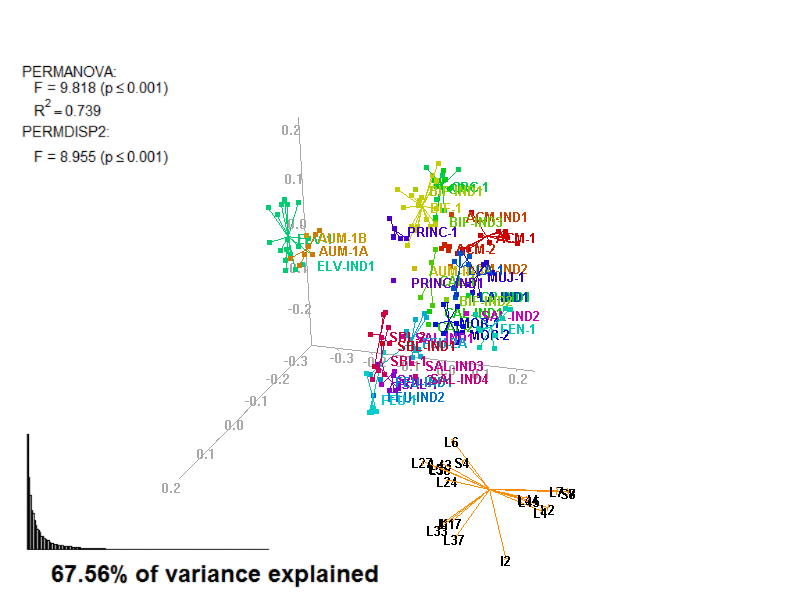
Prot2a_Biplot3D.gif
biplot2d3d::biplot_3d(prot2b_3d,
ordination_method = "NMDS",
point_type = "point",
groups = factor_list$FabricGroup,
group_color = color_list$FabricGroup,
group_representation = "stars",
star_centroid_radius = 0,
star_label_cex = .8,
arrow_min_dist = .5,
arrow_body_length = .025,
subtitle = prot2b_3d$sub3D,
test_text = prot2b_tests$text(prot2b_tests),
test_cex = 1.25,
test_fig = c(0, 0.5, 0.65, .99),
view_zoom = 0.9)
biplot2d3d::animation(directory = directories$prot2,
file_name = "Prot2b_Biplot3D")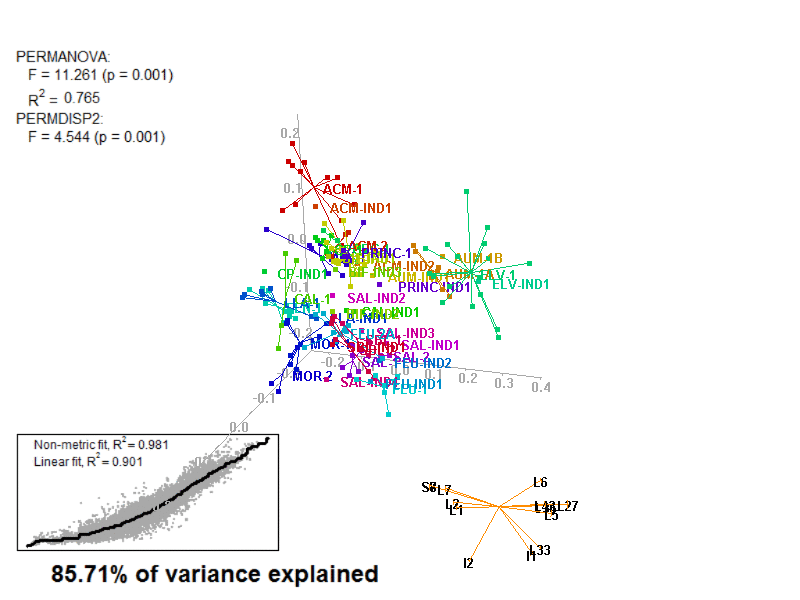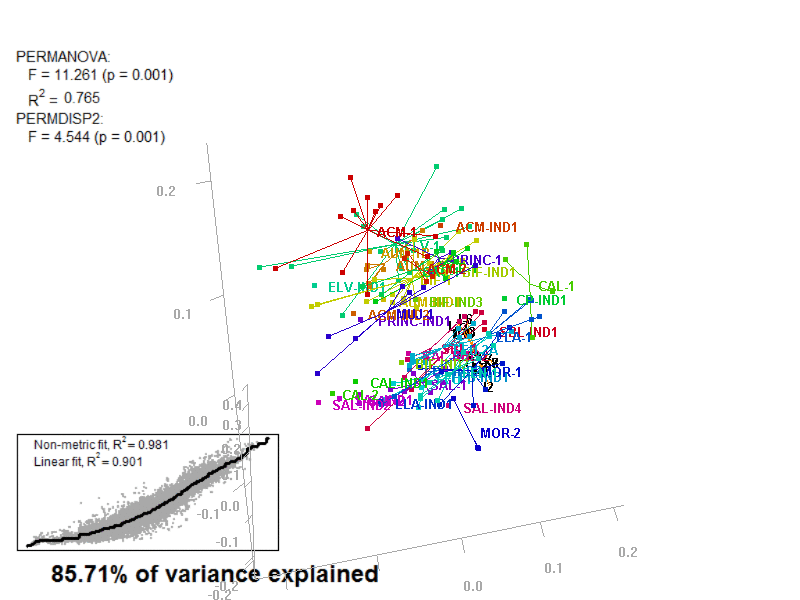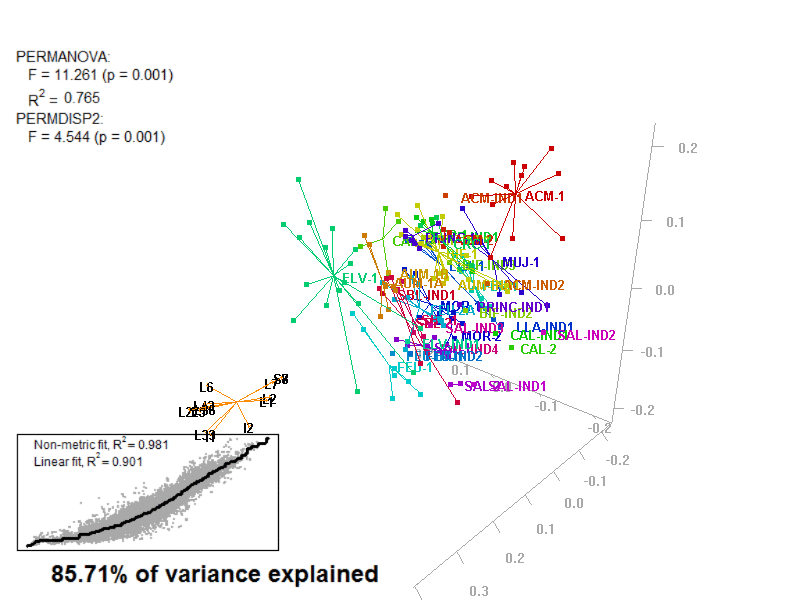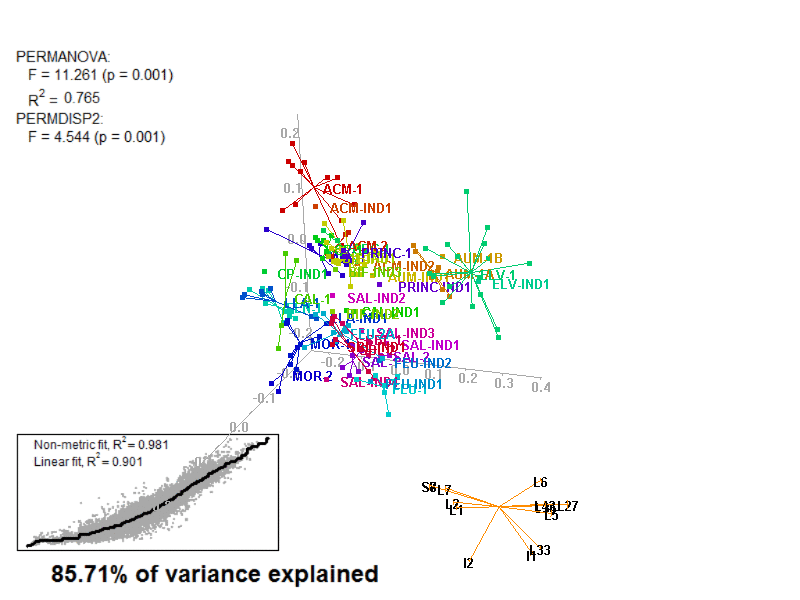
Prot2b_Biplot3D_snapshot.png